MR-KG: A Knowledge Graph of Mendelian Randomization Evidence Powered by Large Language Models
📌 Background
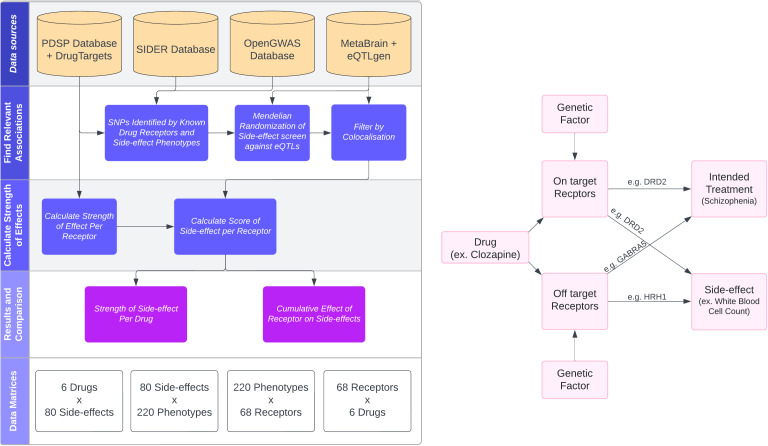
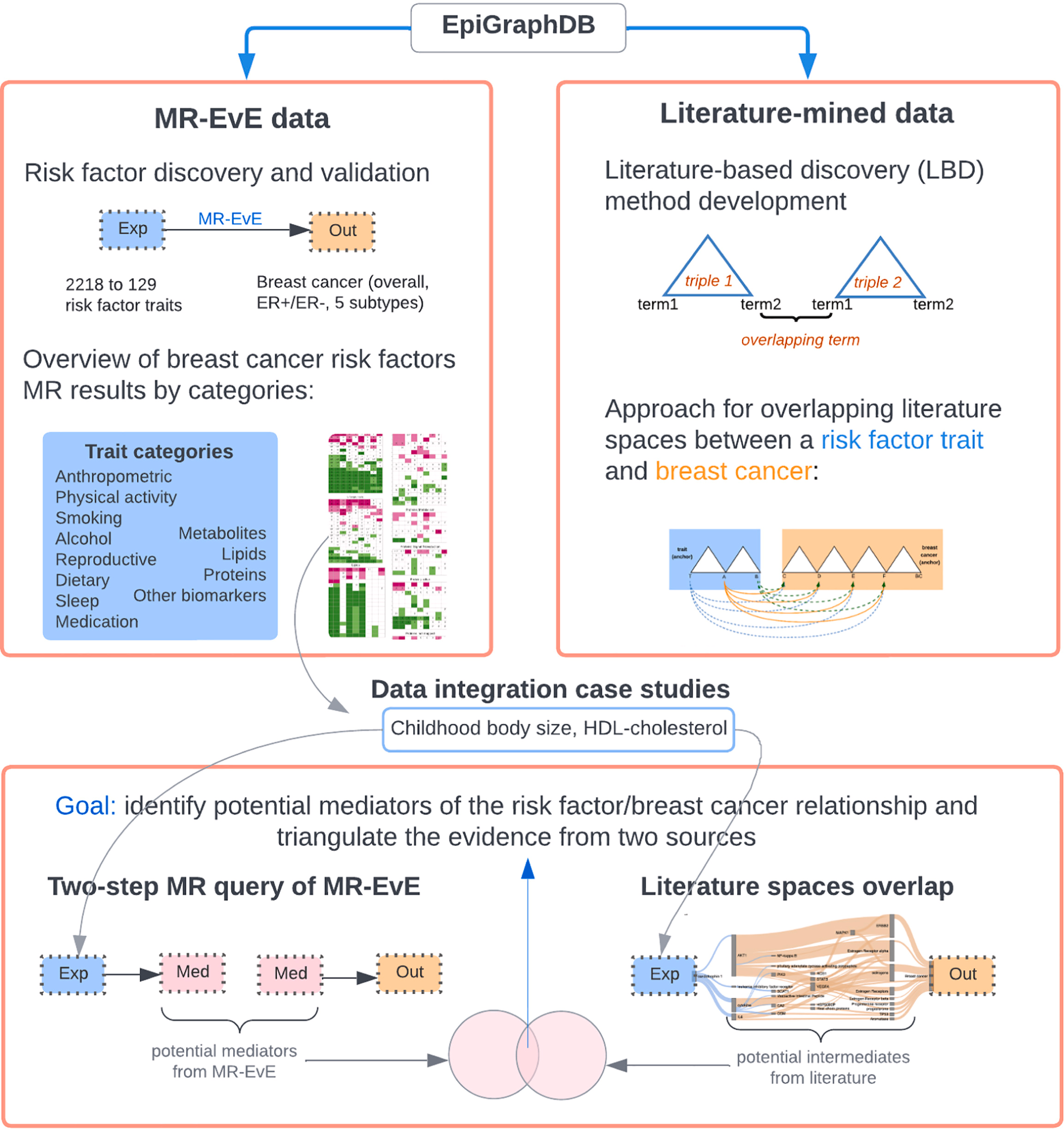
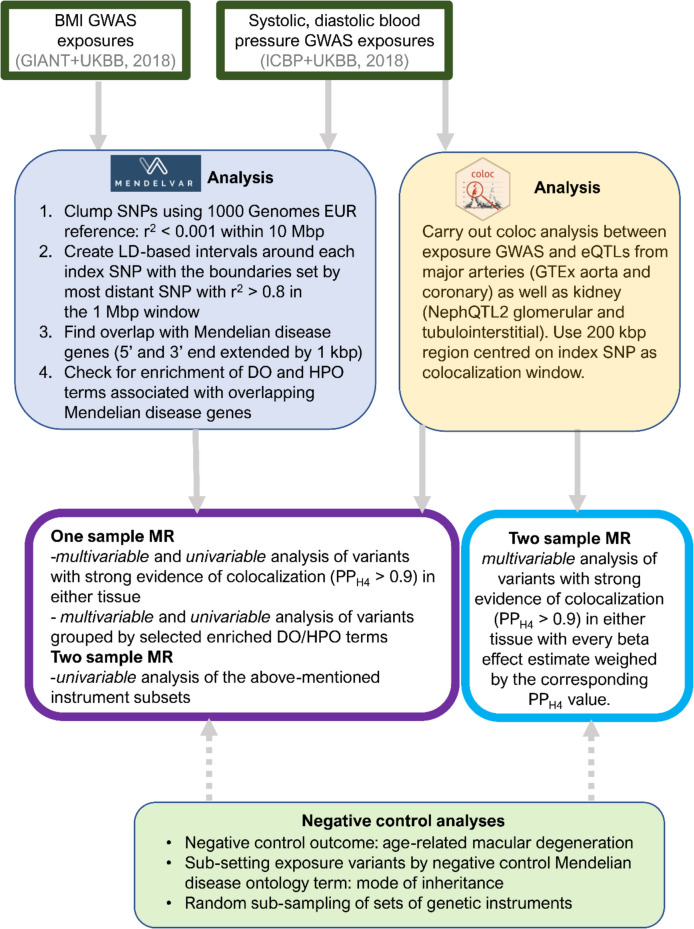
Mendelian randomization (MR) is a powerful causal inference method that uses genetic variants as natural experiments to assess causal relationships between putative risk factors and disease outcomes. MR studies are increasingly abundant, but synthesising evidence across them remains challenging due to heterogeneity in reporting, traits examined, and the structure of the published literature.
To address this, Liu, Burton, Gatua, Hemani & Gaunt (2025) introduce MR-KG — a knowledge graph of MR evidence automatically extracted from published studies using large language models (LLMs).