Integrating Mendelian randomization and literature mining to map breast cancer risk factors
Breast cancer research spans epidemiology, molecular biology, clinical trials, and a vast and rapidly growing literature. One challenge is triangulating across these evidence types: when different sources point in the same direction, we can be more confident we are seeing something causal rather than correlational.
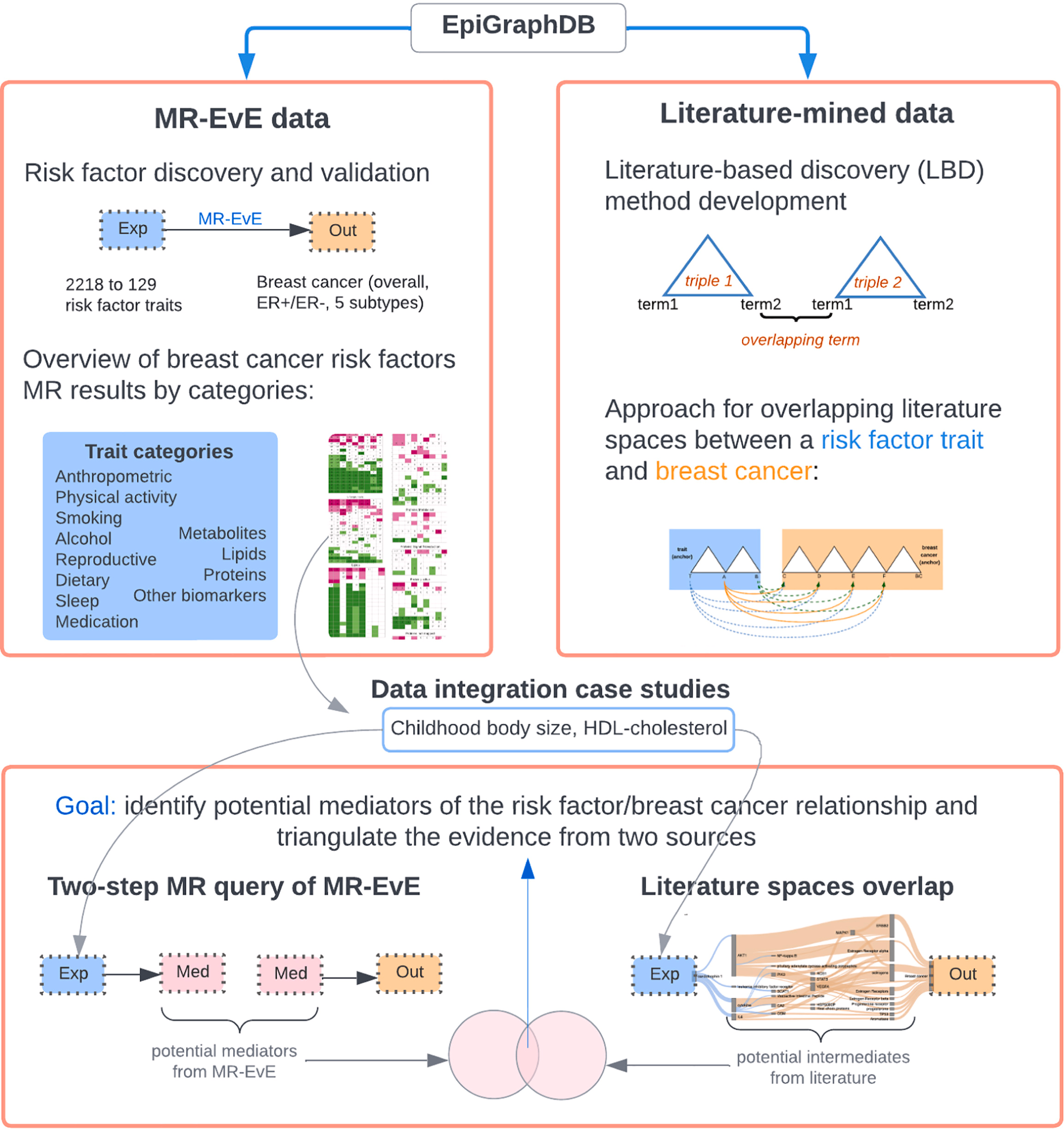
In a paper led by Marina Vabistsevits published in the Journal of Biomedical Informatics, we show how to bring two complementary sources together:
- Mendelian randomization (MR) evidence generated at scale using MR-EvE (“Everything-vs-Everything”), and
- Literature-mined relationships stored in EpiGraphDB, our biomedical knowledge graph.
Why combine MR with literature mining?
MR can help prioritise likely causal risk factors, but it does not automatically tell us how an exposure influences disease. Meanwhile, the biomedical literature is full of mechanistic clues—but it is too large to read manually, and individual papers can be hard to weigh.
Our aim was to use MR for efficient hypothesis generation, and then use literature-mined links to suggest plausible intermediates/mediators, before returning to genetics again for validation.
What we did
We started with MR-EvE estimates to screen many traits against breast cancer outcomes, looking for candidate risk factors and possible mediators. We then integrated these MR results with literature-mined “triples” (subject–predicate–object statements extracted from papers) in EpiGraphDB, using an approach based on overlapping “literature spaces” between a risk factor trait and breast cancer.
Finally, for literature-based discovery (LBD) candidates, we used two-step MR to check whether a proposed intermediate sat on a plausible causal path from risk factor → intermediate → breast cancer.
What we found
Using this pipeline, we identified 129 lifestyle risk factors and molecular traits with evidence of an effect on breast cancer (including both established and potentially novel signals). We also made the MR results explorable via an R/Shiny app for interactive browsing and hypothesis generation.
To show how the integration works in practice, the paper walks through two case studies:
- Childhood body size, where combining MR and literature helps explore downstream intermediates that might connect early-life adiposity to later breast cancer risk.
- HDL-cholesterol, where the literature-mined links provide mechanistic hypotheses that can then be followed up using genetics-based mediation checks.
Why this matters
This is not about replacing careful study design or detailed mechanistic work. The point is to make it easier to navigate the space of plausible hypotheses, and to prioritise follow-up work with a clearer view of (a) what looks causal and (b) what the literature suggests about potential pathways.
More broadly, it’s a demonstration of what we think knowledge graphs are good at: connecting evidence across study types and helping us ask better questions, faster.
Try it yourself
- Paper: Integrating Mendelian randomization and literature-mined evidence for breast cancer risk factors (Journal of Biomedical Informatics, 2025).
- Interactive MR heatmaps app: https://mvab.shinyapps.io/MR_heatmaps/
- EpiGraphDB platform: https://epigraphdb.org
If you use the app or EpiGraphDB in your work and have ideas for additional features, do get in touch — we’re always keen to hear how people are using these resources.