Dissecting blood pressure and BMI a pathway- and tissue-partitioned Mendelian randomization comparison
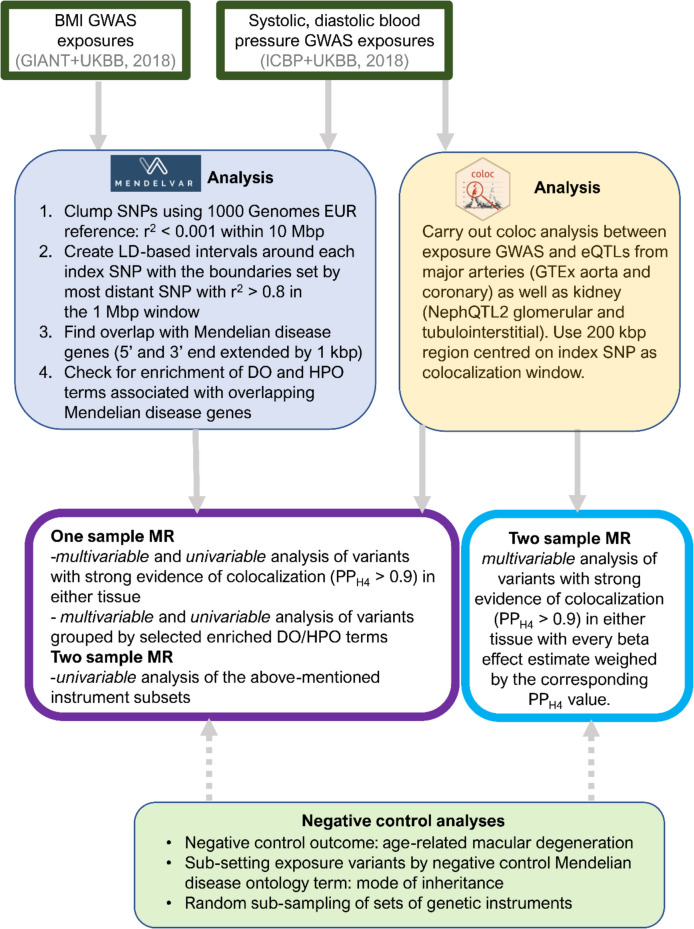
Complex traits like blood pressure (BP) and body mass index (BMI) are highly polygenic: hundreds of associated variants can be used as instruments in Mendelian randomization (MR). But those variants don’t all “mean the same thing” biologically—some may act through kidney physiology, others through vasculature, neurobiology, metabolism, and so on. If we can separate instruments into interpretable biological subsets, we can start asking questions like:
- Which component of BP is most responsible for coronary heart disease risk?
- Are BMI → atrial fibrillation effects more “metabolic” or more “neuro-behavioural”?
Work led by Genevieve Leyden and Maria Sobczyk and now published in Genome Medicine sets out to do exactly this by comparing two ways of partitioning genetic instruments before running MR.
Two complementary ways to split instruments
The paper evaluates two strategies side-by-side:
1) Pathway-partitioned instruments from Mendelian disease phenotypes
We introduce a new approach that groups genome-wide significant BP/BMI variants by whether they lie near Mendelian disease genes enriched for particular symptom categories (via MendelVar).
- For blood pressure, variants are partitioned into renal vs vessel (vasculature) pathways.
- For BMI, variants are partitioned into metabolic vs mental health pathways.
The idea is pragmatic: Mendelian disorders provide a clinically grounded “phenotype dictionary” that can help annotate the biology sitting underneath GWAS loci.
2) Tissue-partitioned instruments from genetic colocalization
We compare the above to an existing, colocalization-derived approach previously published by Genevieve that partitions variants based on evidence that a GWAS signal overlaps an eQTL signal in specific tissues.
- For blood pressure, variants are assigned to kidney vs artery tissues.
- For BMI, variants are assigned to brain vs adipose tissues.
These two partitioning schemes are not expected to be identical—one is anchored in clinical phenotype enrichment, the other in molecular regulatory context—but comparing them can highlight where biology is robust versus where interpretation needs caution.
What did we find?
Our headline result is that different partitions can suggest different “drivers” of the same exposure–outcome relationship.
Blood pressure: vessel pathways stand out for heart disease, but tissue signals are more nuanced
Using the pathway-partitioned approach, the systolic BP → heart disease effect appeared more strongly driven by vessel (vasculature) instruments than renal instruments.
However, in the tissue-partitioned analyses, the corresponding comparison suggested a stronger effect attributed to kidney than artery tissue, consistent with BP acting through multiple intertwined mechanisms rather than a single clean partition.
Across outcomes, we also report consistent evidence that:
- Vessel (pathway) and artery (tissue) instruments dominate the negative directional effect of diastolic BP on left ventricular stroke volume, and the positive directional effect of systolic BP on type 2 diabetes.
BMI and atrial fibrillation: metabolic vs brain components
When focusing on BMI → atrial fibrillation, we found the causal effect was predominantly driven by:
- metabolic-pathway instruments (relative to mental health pathway instruments), and
- brain-tissue instruments (relative to adipose tissue instruments).
That contrast is interesting in itself: it suggests that “metabolic” and “brain” are not mutually exclusive stories, and may instead reflect overlapping causal routes (e.g. appetite regulation and energy balance).
Why this matters for MR interpretation
A common workflow in MR is to build a single instrument set and estimate an “average” causal effect. This paper is a reminder that, for many complex traits, that average may be combining multiple biological components.
Partitioning instruments can help us:
- identify potential pleiotropic pathways (e.g. if only one partition shows an effect),
- generate mechanism-focused hypotheses (which component of BP/BMI is most relevant?),
- prioritize follow-up (e.g. deeper locus-to-gene work in the partition driving an effect), and
- spot interpretability pitfalls when different annotation schemes tell different stories.
However, we emphasize that partitioned effect-size differences need robust validation (and can arise by chance without strong supporting evidence), so this should be viewed as a hypothesis-generating framework rather than a final answer.
Our take
From our perspective, this is a neat example of how we can bring in external biological knowledge—here, Mendelian disease phenotype enrichment—to get more out of standard GWAS-derived instruments, and to complement more “molecular” approaches like colocalization.
If you are doing MR on a complex exposure and you suspect heterogeneous mechanisms, it’s worth thinking about instrument partitioning early: it can make your results easier to interpret, and it can help decide what’s worth chasing downstream.
Paper
Leyden GM, Sobczyk MK, Richardson TG, Gaunt TR. Distinct pathway-based effects of blood pressure and body mass index on cardiovascular traits: comparison of novel Mendelian randomization approaches. Genome Medicine (2025) 17:54. DOI: 10.1186/s13073-025-01472-2. PubMed: https://pubmed.ncbi.nlm.nih.gov/40375348/. Full text: https://pmc.ncbi.nlm.nih.gov/articles/PMC12079859/