Genetics as a side‑effect detective for antipsychotic medicines
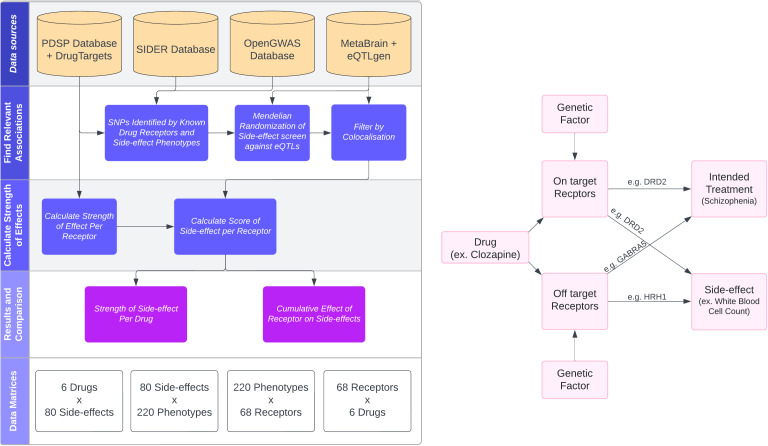
Side‑effects are one of the main reasons people stop taking antipsychotic medicines — even when the drugs are helping with symptoms. But when someone reports “I’ve gained weight” or “my blood pressure has changed”, it’s often hard to know whether the drug truly caused it, which biological target is responsible, and whether that target is the one we wanted to hit in the first place.
In work led by Andrew Elmore, published in PLOS Genetics, we combine pharmacology (what receptors a drug binds) with human genetics (natural experiments) to map side‑effects back to specific receptors.
The basic idea
Antipsychotics don’t just bind a single receptor. They bind many — and some of those bindings are “on‑target” (part of how the drug works), while others are “off‑target” (biological collateral damage).
We built a framework that brings together:
- Drug–receptor binding affinities (how strongly each drug binds each receptor)
- Reported side‑effects from a large reference database
- Genetic instruments for receptor/gene activity (eQTLs)
- GWAS traits that can stand in for side‑effects
- Mendelian randomization (MR) + genetic colocalisation to strengthen causal interpretation
The output is a simple summary: a side‑effect score for each Drug × Receptor × Trait combination, which we can then aggregate to compare drugs and mechanisms.
What we analysed
We focused on six commonly prescribed antipsychotics (including clozapine, olanzapine and risperidone). Across these drugs we identified 68 receptors with evidence of binding, and started from 165 reported side‑effects — of which 80 could be genetically proxied using available GWAS in OpenGWAS.
What we found
A few results stood out:
- We identified 36 side‑effects that look likely to be caused by drug action through 30 receptors.
- The bulk of evidence pointed to off‑target mechanisms.
- Clozapine showed the largest cumulative side‑effect profile and the largest number of scored side‑effects in this framework.
Three concrete examples
The paper walks through three side‑effects in detail (and these nicely illustrate how the approach can generate mechanistic hypotheses).
1) Neutropenia (dangerously low white blood cell counts)
Clozapine is clinically linked to rare but serious neutropenia. In our genetic scoring, the signal for neutropenia was strongest for clozapine and suggested contributions from targets including GABRA1 and HTR1B.
2) Weight gain
Weight gain is a major concern for patients and a common reason for discontinuation. Our results again highlighted clozapine (and olanzapine) and suggested that differences in binding to targets such as CHRM3 and HRH1 could help explain why these drugs tend to have larger weight‑gain profiles than others.
3) Blood pressure effects
Clinical evidence on antipsychotics and blood pressure can be mixed, and we saw that different receptors imply different directions of effect. In the scoring, blood‑pressure signals were strongly influenced by HRH1, consistent with this receptor being a plausible driver for some blood‑pressure changes.
Why this matters
What we think is important about this work is the transferability:
- It provides a framework to triage side‑effects early (even before large trials, when we have target information and genetics).
- It can help separate “likely causal” side‑effects from those that may be coincidental, comorbidity‑driven, or reporting artefacts.
- It gives mechanistic handles: if a side‑effect seems driven by an off‑target receptor, that receptor becomes a candidate to avoid in future drug design.
This won’t replace clinical pharmacovigilance. But it could help researchers ask better questions — and focus lab, trial, and monitoring efforts where the biology is most convincing.
Caveats we should keep in mind
A few limitations are worth emphasising:
- Some receptor families (notably parts of the GABA system) have incomplete binding‑affinity data, which affects how confidently we can compare magnitudes across drugs.
- The genetic resources used are predominantly European‑ancestry, so we should be careful about generalising across populations.
- Pharmacokinetics (dose, tissue penetration, blood–brain barrier) and real‑world reporting can complicate “genetics → drug” translation.
Links
- Paper (open access): https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1011793
- PubMed record: https://pubmed.ncbi.nlm.nih.gov/40720497/
- Code example (linked from the paper): https://github.com/andrew-e/side-effect-score