Overview
The BioRxiv and MedRxiv preprint facilities are vital infrastructure for the biomedical research community, which also provide a rich and comprehensive resource for data mining biomedical literature for investigations on research trends, interests, and novel findings. In our previous works we have conducted extensive literature mining efforts on BioRxiv and MedRxiv to extract structural literature knowledge into EpiGraphDB[1] and derive research claims from recent preprints to be triangulated with other evidence on ASQ[2].
Supported by the Elizabeth Blackwell Institute Rapid Research Funding Call, in this project we have acquired the full text data archives for BioRxiv and MedRxiv preprints from 2013 to May 2023, and we have also conducted some exploratory analysis on the data archives.
Full text archives of BioRxiv / MedRxiv preprints
Web scraping pre-print text is time consuming as well as error prone. However, BioRxiv and MedRxiv have provided archive data on the preprints for the purpose of text and data mining hosted on Amazon AWS S3 as Requester Pays Buckets. Full text archives from BioRxiv and MedRxiv are stored in two S3 buckets biorxiv-src-monthly and medrxiv-src-monthly respectively, where the organization structure is roughly as follows (high level description is also available on the tdm pages):
biorxiv-src-montly
├── Back_Content
└── Current_Content
├── April_2019
├── April_2020
│ ├── 0002415e-6e79-1014-bad3-d7b11ff8718c.meca
│ ├── 0008729e-7222-1014-9e12-b08e9cbb4568.meca
│ ├── 00114dfb-6f21-1014-b58f-80aa2e0e89bd.meca
│ ├── 00114e62-6cf9-1014-aedd-beed0b185e0b.meca
│ ├── 0025be66-6dea-1014-8d12-849fadd63f55.meca
│ ├── ....
│ ├── ....
├── April_2021
├── April_2022
├── April_2023
├── August_2019
├── August_2020
├── August_2021
├── August_2022
├── December_2018
├── December_2019
├── December_2020
├── December_2021
├── December_2022
├── ...
├── ...
medrxiv-src-monthly
├── Back_Content
└── Current_ContentEach .meca file is an archive in .zip format which contains individual files associated with one preprint submission. For example the following structure corresponds to files associated with this preprint (Shrestha et al., 2022, Knowledge, Attitude and Practice (KAP) study on COVID-19 among the general population of Nepal). Specifically the file ./content/22279527.xml is the “full text / manuscript” file in .xml format.
0a2ef310-6c04-1014-8ee5-ac250845df11.meca
├── content
│ ├── 22279527.pdf
│ ├── 22279527v1_tbl1.tif
│ ├── 22279527v1_tbl2.tif
│ ├── 22279527v1_tbl3.tif
│ ├── 22279527v1_tbl4.tif
│ ├── 22279527v1_tbl5a.tif
│ ├── 22279527v1_tbl5.tif
│ ├── 22279527v1_tbl6a.tif
│ ├── 22279527v1_tbl6b.tif
│ ├── 22279527v1_tbl6.tif
│ └── 22279527.xml
├── directives.xml
├── manifest.xml
├── mimetype
└── transfer.xmlHow do we know which .meca archive filename corresponds to which preprint DOI? Unfortunately we haven’t found a way to know this without actually opening the meca file and parsing the full text xml. In addition we have identified several rounds of changes to the storage structure, as well as text format (e.g. how a key information such as submission date is represented in xml tags), which means that it takes effort to systematically and robustly curate various information such as metadata as well as sections (e.g. abstracts, methods, results, conclusions) in the manuscripts. We will need to curate these information as separate intermediate datasets for our follow-up research projects.
Some exploratory results
As part of our on-going efforts in parsing and curating the preprint information from the raw archives, here we discuss some of the gathered results which will share insights into future projects.
Trends and distributions
Figures 1-3 shows the trends and distributions of the preprints. MedRxiv was separated from BioRxiv in 2019 to host preprints related to clinical and medical topics, and they have now become vital infrastructure to the scientific community in communicating rapid findings. The COVID-19 pandemic also substantially contributed to this growth in researchers presenting their findings as preprints before the peer-review process is complete. Bioinformatics, Cancer Biology, Cell Biology, Evolutionary Biology, Microbiology, as well as Neuroscience are among the most populous categories in BioRxiv, whereas Epidemiology, Infectious Diseases (except HIV/AIDS), Public and Global Health are among the most populous ones in MedRxiv.
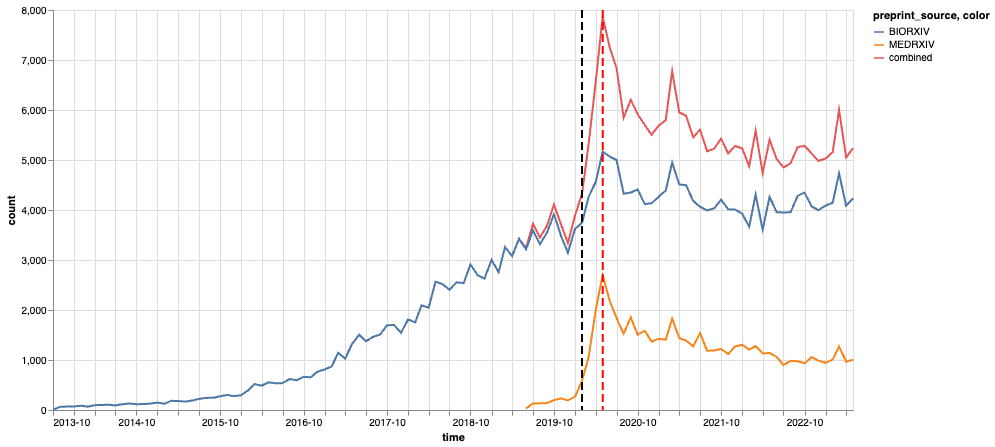 Figure 1: Total number of monthly preprint submissions We count the archives by their submission month and preprint source (biorxiv / medrxiv). The red line corresponds to the combined submissions from both sources. The black dashed line corresponds to February 2020 when COVID-19 started to a pandemic, and the red dash line corresponds to May 2020 around which the monthly submissions peaked.
Figure 1: Total number of monthly preprint submissions We count the archives by their submission month and preprint source (biorxiv / medrxiv). The red line corresponds to the combined submissions from both sources. The black dashed line corresponds to February 2020 when COVID-19 started to a pandemic, and the red dash line corresponds to May 2020 around which the monthly submissions peaked.
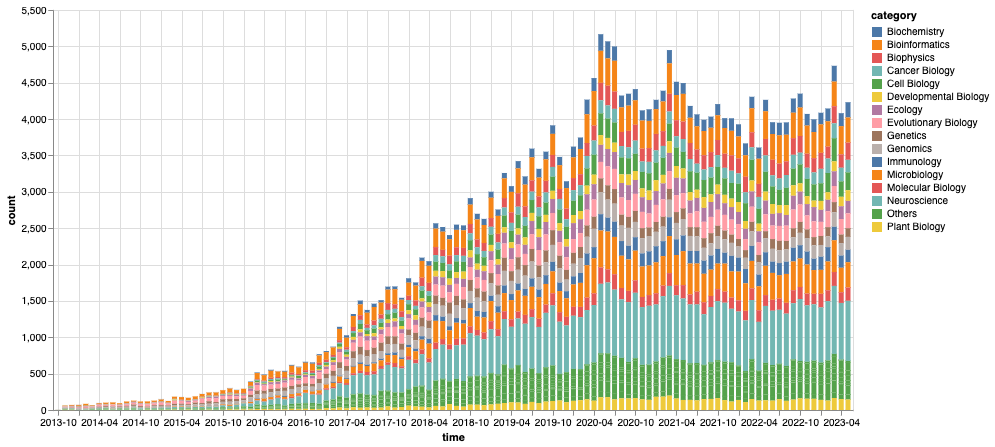 Figure 2: Total number of montly BioRxiv submissions by category We count the archives by their submission month and category, where we consolidate the monthly categories below top-15 most populous ones into an “Others” category.
Figure 2: Total number of montly BioRxiv submissions by category We count the archives by their submission month and category, where we consolidate the monthly categories below top-15 most populous ones into an “Others” category.
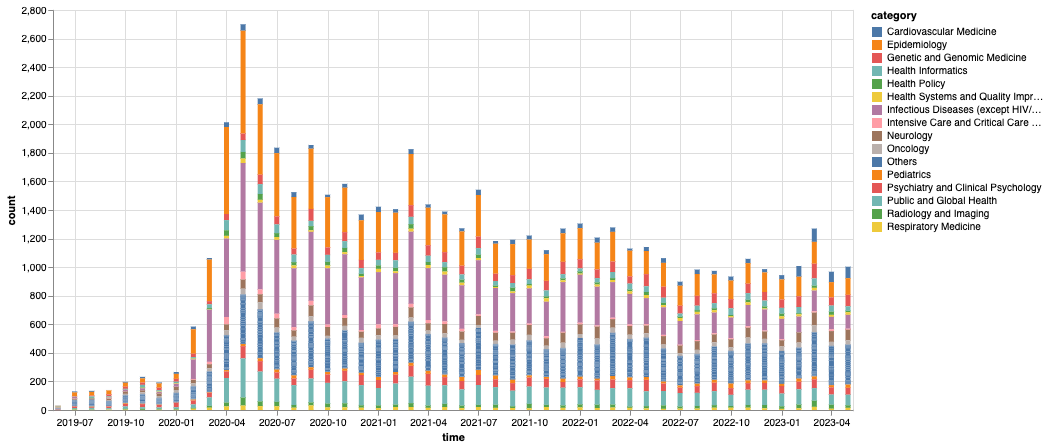 Figure 3: Total number of montly MedRxiv submissions by category We count the archives by their submission month and category, where we consolidate the monthly categories below top-15 most populous ones into an “Others” category.
Figure 3: Total number of montly MedRxiv submissions by category We count the archives by their submission month and category, where we consolidate the monthly categories below top-15 most populous ones into an “Others” category.
Topic analysis
To further demonstrate the distribution of research topics, we did some simple topic analysis, where we used the BERTopic library to efficiently embed and model all (over 300,000) article titles to derive topics corresponding to research topics. Figure 4 shows the trends of the dynamic topics, where we visually identify a topic related to the transmission of SARS-CoV-2 virus (“sarscov2”, “transmission”, “seroprevalence”, etc.) dominate over others during the early phase (2020-2021) of the pandemic and its gradual decline. In addition, a topic related to vaccines (“vaccination”, “vaccine hesitancy”, etc.) can also been seen being prevalent during 2021. Figure 5 then further demonstrates how each topic relate to others in the topic clusters.
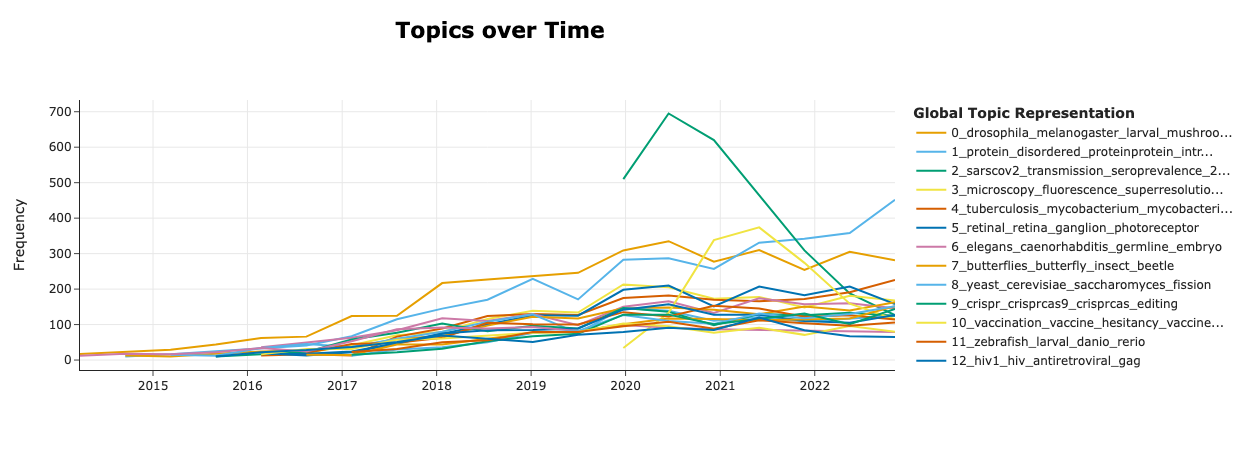 Figure 4: Trends of topics dynamic topic modelling analysis Dynamic topics are modelled by associating the submission month of the preprint with the topics represented from the article titles.
Figure 4: Trends of topics dynamic topic modelling analysis Dynamic topics are modelled by associating the submission month of the preprint with the topics represented from the article titles.
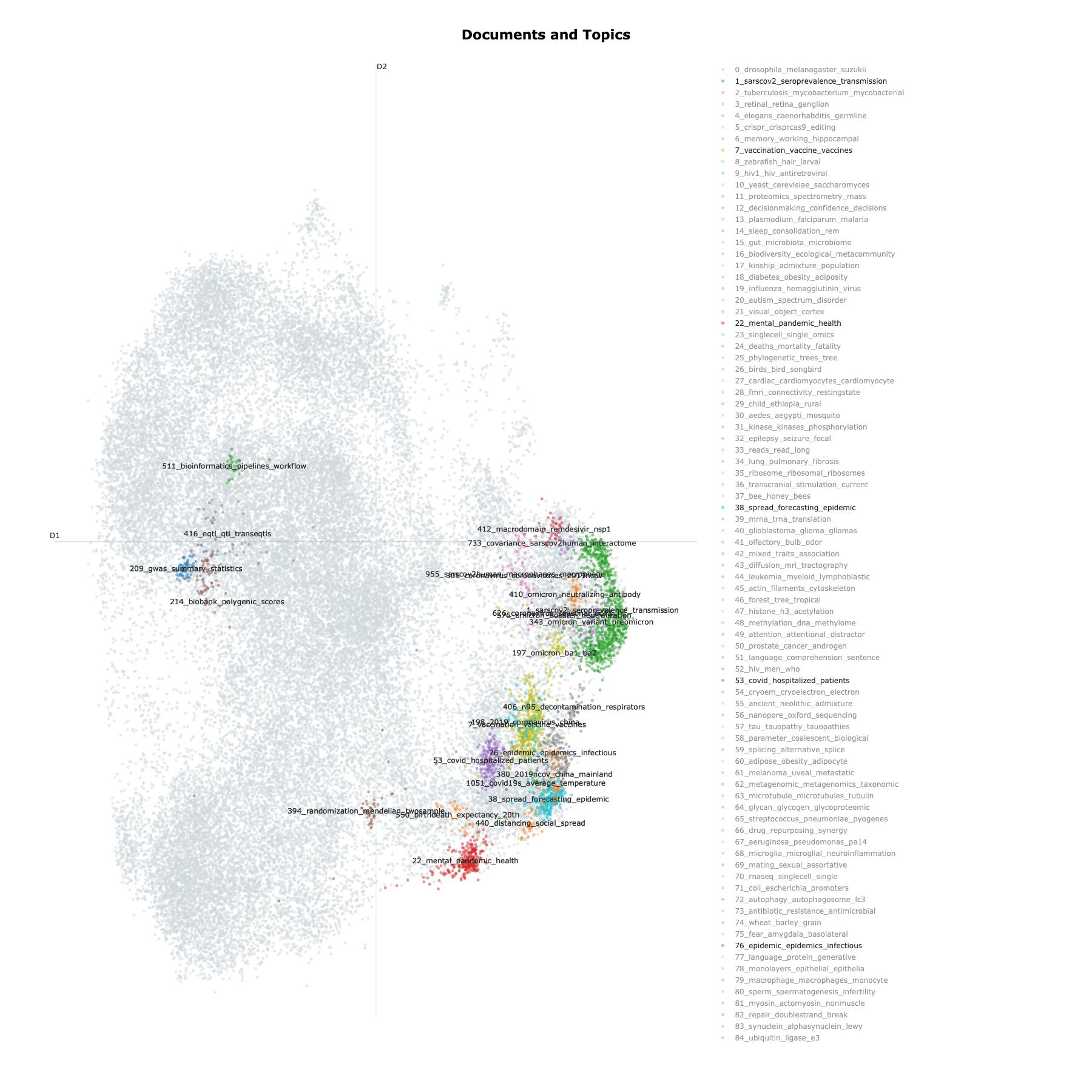 Figure 5: Topic clusters We randomly select a sample of 100_000 documents (article titles) to generate the topic model, and then used the UMAP clustering method to visualize the clustering of documents. For all the generated topics, we highlight those associated with the COVID-19 pandemic, as well as for comparison a few others related to health data science and genetic epidemiology topics.
Figure 5: Topic clusters We randomly select a sample of 100_000 documents (article titles) to generate the topic model, and then used the UMAP clustering method to visualize the clustering of documents. For all the generated topics, we highlight those associated with the COVID-19 pandemic, as well as for comparison a few others related to health data science and genetic epidemiology topics.
Summary
The above preliminary results on the exploratory analysis demonstrate the richness in what can be data mined from analysing the BioRxiv / MedRxiv preprints. We are excited at the prospect of research projects on text mining biomedical studies involving the full historical archive of biomedical research (as proxied by the preprints), not just on public health and epidemiology topics, but also cross-discipline ones such as social-economic factors influencing the eventual publication of a preprint, effective modelling of research trends via various network or clustering analysis methods. For potential collaboration interests please reach out to Yi Liu (yi6240.liu[at]bristol.ac.uk) and Tom Gaunt (tom.gaunt[at]bristol.ac.uk).
Code availability
Our code in data accessing as well as research analysis have been published as a GitHub repository here https://github.com/MRCIEU/biorxiv-medrxiv-tdm, which we will continue to update in the future as our current results are just a first step towards bigger analysis projects.
References
[1] Yi Liu, Benjamin Elsworth, et al., Tom Gaunt, EpiGraphDB: a database and data mining platform for health data science, Bioinformatics, Volume 37, Issue 9, May 2021, Pages 1304–1311, https://doi.org/10.1093/bioinformatics/btaa961
[2] Yi Liu, Tom R Gaunt, Triangulating evidence in health sciences with Annotated Semantic Queries medRxiv 2022.04.12.22273803; doi: https://doi.org/10.1101/2022.04.12.22273803