Overview
Genetic studies have been very biased towards populations of European ancestry in western Europe and the United States of America, and this has led to a significant bias in the application of Mendelian randomization (MR) to identify intervention targets. In this project we worked with a leading international genetics consortium, the Global Biobank Meta-analysis Initiative (GBMI) to evaluate the differences in predicted drug target effects between African and European ancestry populations.
What we did
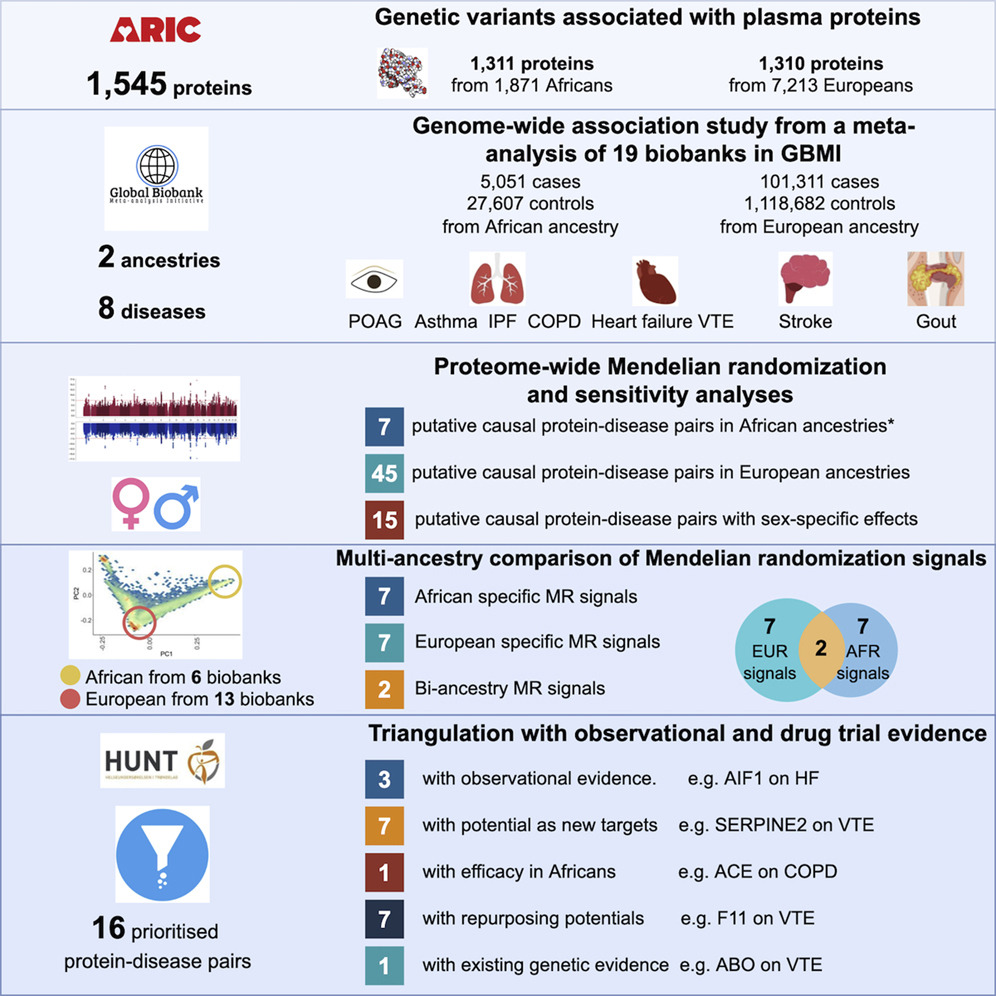
In this paper (published in Cell Genomics), Huiling and Chris carried out a multi-ancestry proteome-wide MR analysis using cross-population data from GBMI. We estimated the causal effects of 1545 proteins on eight diseases in African (n=32,658) and European (n=1,219,993) ancestry populations. We found 45 protein-disease pairs with MR and colocalization evidence of causality in European ancestry, and 7 protein-disease pairs with evidence of causality in African ancestry. The difference in sample size (and, consequently, statistical power) almost certainly explains the large difference in number of causal effects detected. Interestingly, only 2 protein-disease pairs showed MR evidence in both ancestries.
Paper
‘Proteome-wide Mendelian randomization in global biobank meta-analysis reveals multi-ancestry drug targets for common diseases’ by Huiling Zhao, Humaria Rasheed, Therese Haugdahl Nøst, Yoonsu Cho, Yi Liu, Laxmi Bhatta, Arjun Bhattacharya; Global Biobank Meta-analysis Initiative; Gibran Hemani, George Davey Smith, Ben Michael Brumpton, Wei Zhou, Benjamin M Neale, Tom R Gaunt and Jie Zheng in Cell Genom. 2022 Nov 9;2(11):None. doi: 10.1016/j.xgen.2022.100195..