Overview
Molecular quantitative trait loci (molQTL), which can provide functional evidence on the mechanisms underlying phenotype-genotype associations, are increasingly used in drug target validation and safety assessment. In particular, protein abundance QTLs (pQTLs) and gene expression QTLs (eQTLs) are the most commonly used for this purpose. However, questions remain on how to best consolidate results from pQTLs and eQTLs for target validation.
What we did
In this bioRxiv pre-print we combined blood cell-derived eQTLs and plasma-derived pQTLs to form QTL pairs representing each gene and its product. We performed a series of enrichment analyses to identify features of QTL pairs that provide consistent evidence for drug targets based on the concordance of the direction of effect of the pQTL and eQTL. We repeated these analyses using eQŢLs derived in 49 tissues.
We found that 25-30% of blood-cell derived QTL pairs have discordant effects. The difference in tissues of origin for molecular markers contributes to, but is not likely a major source of, this observed discordance. Finally, druggable genes were as likely to have discordant QTL pairs as concordant.
Our analyses suggest combining and consolidating evidence from pQTLs and eQTLs for drug target validation is crucial and should be done whenever possible, as many potential drug targets show discordance between the two molecular phenotypes that could be misleading if only one is considered. We also encourage investigating QTL tissue-specificity in target validation applications to help identify reasons for discordance and emphasise that concordance and discordance of QTL pairs across tissues are both informative in target validation.
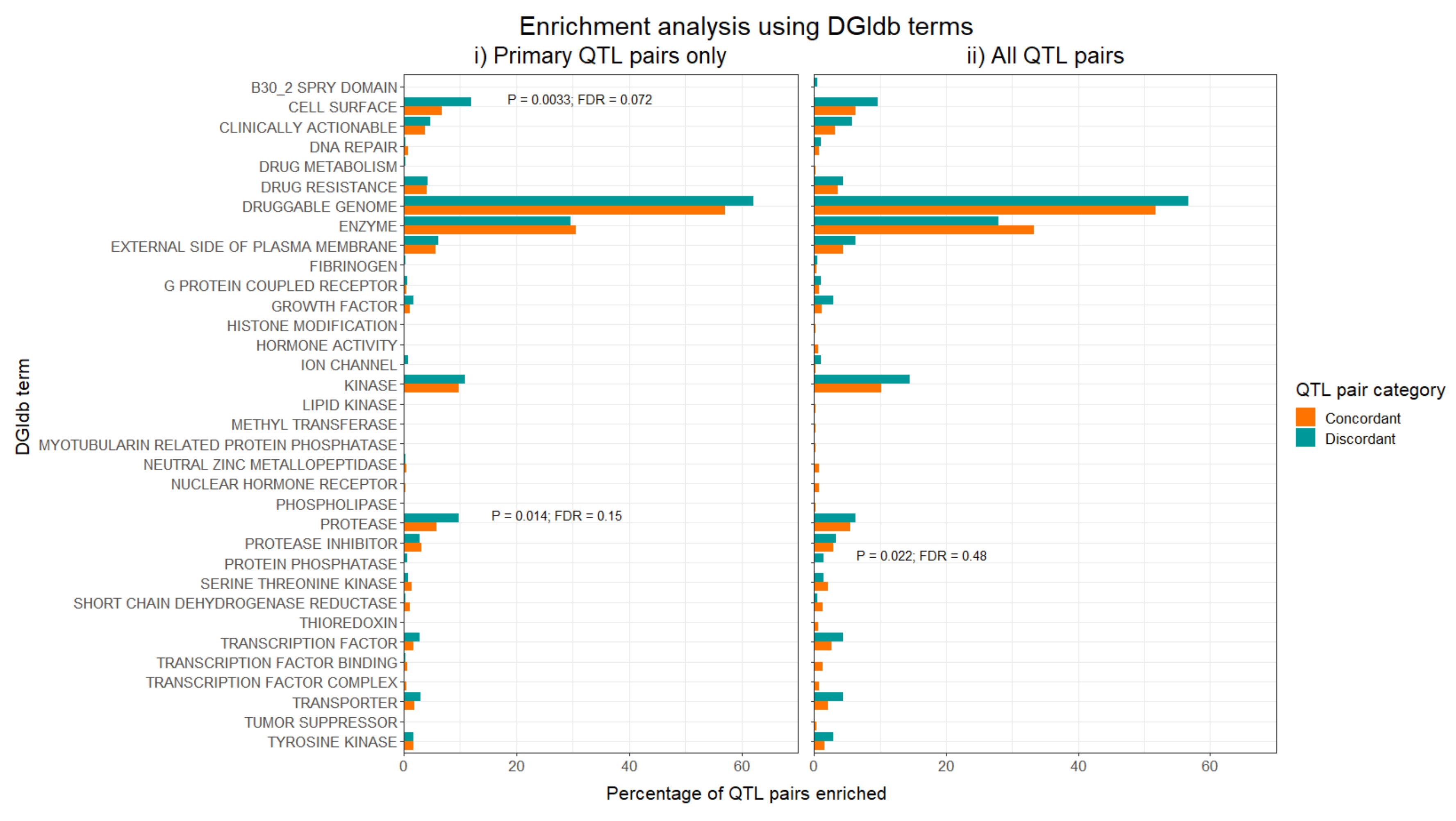 Figure 1: Results for the enrichment analysis using the Drug-Gene Interaction database (DGIdb) for druggability-related terms. The bars show the percentage of concordant or discordant QTL pairs which were enriched for a given term. P values were calculated using Fisher’s exact test, and unadjusted and FDR-adjusted P values are shown for those terms which reached at least nominal significance.
Figure 1: Results for the enrichment analysis using the Drug-Gene Interaction database (DGIdb) for druggability-related terms. The bars show the percentage of concordant or discordant QTL pairs which were enriched for a given term. P values were calculated using Fisher’s exact test, and unadjusted and FDR-adjusted P values are shown for those terms which reached at least nominal significance.
Paper
‘Evaluating the potential benefits and pitfalls of combining protein and expression quantitative trait loci in evidencing drug targets’ by Jamie W Robinson, Thomas Battram, Denis A Baird, Philip C Haycock, Jie Zheng, Gibran Hemani, Chia-Yen Chen and Tom R Gaunt in bioRxiv.