Overview
Integrating information from data sources representing different study designs has the potential to strengthen evidence in population health research. However, this concept of evidence “triangulation” presents a number of challenges for systematically identifying and integrating relevant information.
In this medRxiv preprint we present ASQ (Annotated Semantic Queries), a natural language query interface to the integrated biomedical entities and epidemiological evidence in EpiGraphDB . ASQ enables users to extract “claims” from a piece of unstructured text, and then investigate the evidence that could either support, contradict the claims, or offer additional information to the query.
The ASQ approach has the potential to support the rapid review of pre-prints, grant applications, conference abstracts and articles submitted for peer review. ASQ implements strategies to harmonize biomedical entities in different taxonomies and evidence from different sources, to facilitate evidence triangulation and interpretation.
ASQ is openly available at https://asq.epigraphdb.org.
What we did
The ASQ platform was designed as a natural language interface to the EpiGraphDB biomedical knowledge graph. ASQ considers two primary evidence groups in EpigraphDB:
- Triple and literature evidence, comprising semantic triples derived from the biomedical literature
- Association evidence, comprising results from genetic correlation, polygenic risk score association and Mendelian randomization
The user interface accepts free text entry (e.g. the abstract of a pre-print or journal article, the summary of a grant application, etc). We then use SemRep [1] to derive “claim triples” (Subject-PREDICATE-Object). The user then selects a triple of interest for analysis.
ASQ maps biomedical entities in the Subject and Object parts of the claim triple to biomedical entities in EpiGraphDB. The system then retrieves evidence from the two evidence categories (above) that link the Subject and Object.
Figure 1 provides an overview of the architecture of the platform.
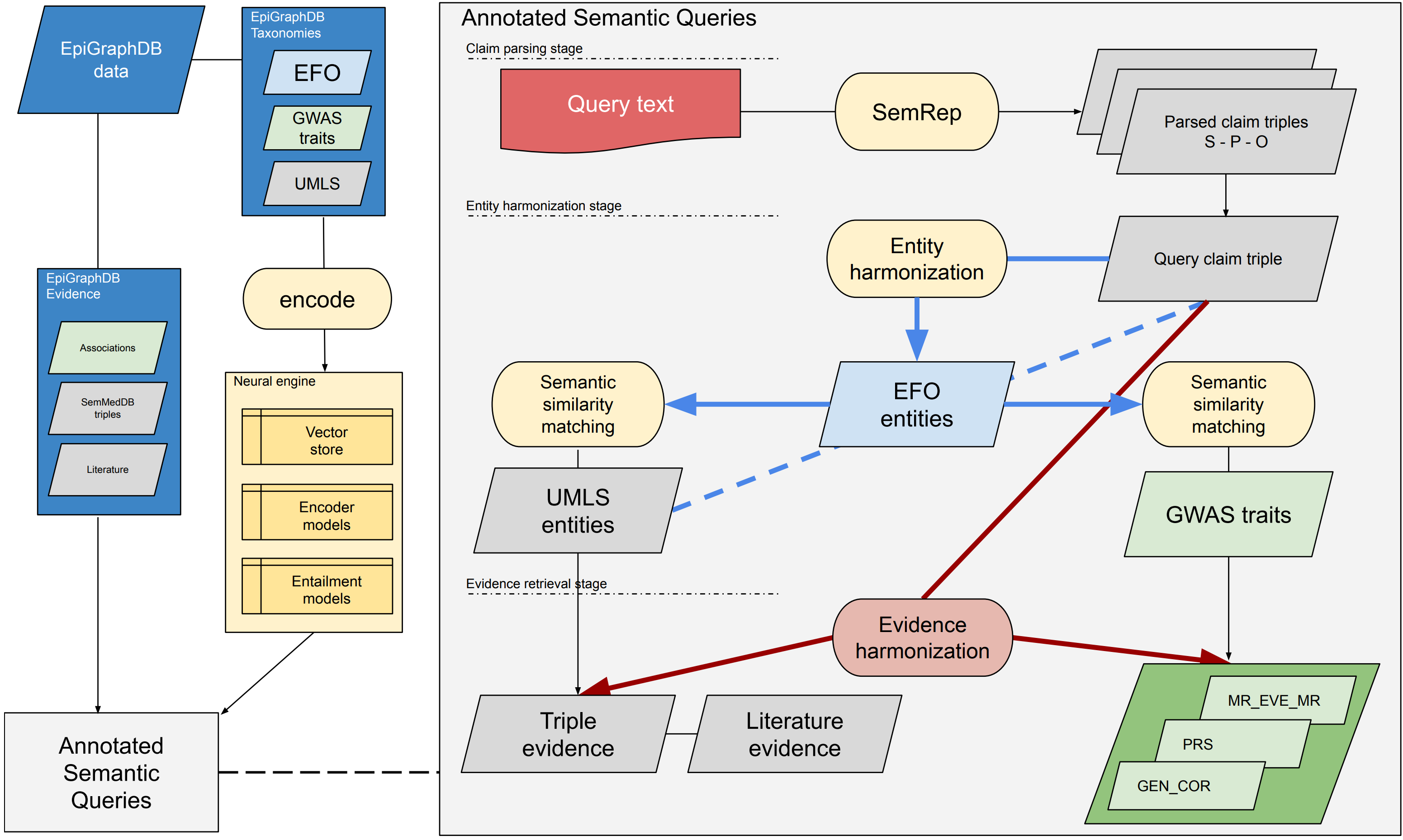 Figure 1: Overall architecture of the EpiGraphDB-ASQ platform Overall architecture design of the EpiGraphDB-ASQ platform and its associated components in the EpiGraphDB ecosystem. Left: EpiGraphDB’s biomedical entities (in the form of graph nodes) from different taxonomies are encoded into vector representations which allows for fast information retrieval against the query of interest. Epidemiological evidence (in the form of graph edges) are incorporated into ASQ as harmonized evidence groups. Right: Internal processing workflow of the EpiGraphDB-ASQ platform by the three stages: the claim parsing stage, the entity harmonization stage, and the evidence retrieval stage
Figure 1: Overall architecture of the EpiGraphDB-ASQ platform Overall architecture design of the EpiGraphDB-ASQ platform and its associated components in the EpiGraphDB ecosystem. Left: EpiGraphDB’s biomedical entities (in the form of graph nodes) from different taxonomies are encoded into vector representations which allows for fast information retrieval against the query of interest. Epidemiological evidence (in the form of graph edges) are incorporated into ASQ as harmonized evidence groups. Right: Internal processing workflow of the EpiGraphDB-ASQ platform by the three stages: the claim parsing stage, the entity harmonization stage, and the evidence retrieval stage
Paper
‘Triangulating evidence in health sciences with Annotated Semantic Queries’ by Yi Liu and Tom Gaunt in medRxiv.
Code availability
Source code for the ASQ platform and relevant analysis scripts can be found via https://github.com/mrcieu/epigraphdb-asq. Tutorial on programmatically accessing the ASQ platform can be found via this Jupyter notebook https://github.com/MRCIEU/epigraphdb-asq/blob/master/analysis/notebooks/programmatic-access.ipynb.
References
[1] Kilicoglu, H., Rosemblat, G., Fiszman, M. & Shin, D. Broad-coverage biomedical relation extraction with SemRep. BMC bioinformatics 21, 1–28 (2020).